From a scientific perspective, 2018 [1] was once again extremely exciting thank to awesome collaborators (at Inria, with DirtyData, and our local scikit-learn team). Rather than going over everything that we did in 2018, I would like to give a few highlights: We published major work using machine learning to map cognition in the brain, We started a new research project on analysis of non-curated data (addressing all of data science, beyond brain imaging); And we worked a lot on growing scikit-learn.
| [1] | It’s already 2019, I am indeed late in posting this summary. |
Highlights
Cognitive brain mapping
We have been exploring how predictive models can help mapping cognition in the human brain. In 2018, these long-running efforts led to important publications.
Atlases of cognition with large-scale human brain mapping
More than 6 years ago, with my student Yannick Schwartz, we started working on compiling an altases of cognition across many cognitive neuroimaging studies. This turned out to be quite challenging for several reasons:
- Formalizing the links between mental processes studied across the literature is challenging. Strictly speaking, every paper studies a different mental process. However, to build an atlas of cognition, we are interested in finding commonalities across the literature.
- While cognitive studies tend to target a specific mental function, the psychological manipulations that they use also recruit many other processes. For instance, a memory study might use a visual n-back task, and hence recruit the visual cortex. The problem is more than an experimental inconvience: varying details of an experiment may trigger different cognitive processes. For instance, there are common and separate pathways for visual word recognition and auditory word recognition.
- Simply detecting regions that are recruited in a given mental operation leads to selecting the whole cortex with enough statistical power. Indeed tasks are never fully balanced; reading might for instance require more attention than listening.
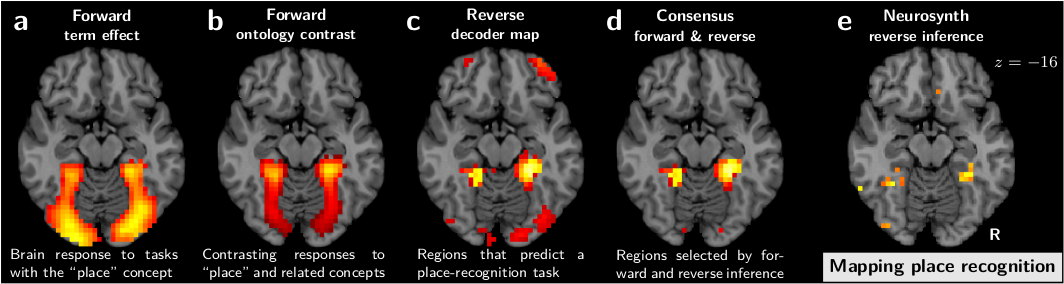
These challenges are related on the one hand to the problem of reverse inference [2], and on the other hand to that of mental-process decomposition, or cognitive subtraction, both central to cognitive neuroimaging. They also call for formal knowledge representation, eg by building ontologies, which is a task harder than it might seem at first glance.
| [2] | In essence, the reverse inference problem arises because in a cognitive brain imaging the observed brain activity is a consequence of the behavior, and not a cause. While a conclusion that activity in a brain structure causes a certain behavior is desirable, it is not directly supported by a cognition neuroimaging experiment. |
In our work [Varoquaux et al, PLOS 2018], we tackled these challenges to build atlases of cognition as follows:
- We assigned to each brain-activity image labels describing the multiple mental processes related to the experimental manipulation
- We used decoding –ie prediction of the cognitive labels from the brain activity– to ground a principled reverse inference interpretation: regions selected indeed do imply the corresponding behavior.
- Regions in the atlas were built of brain structures that both implied the corresponding cognition, and were triggered by it (conditional and marginal link), to ground a strong selectivity:
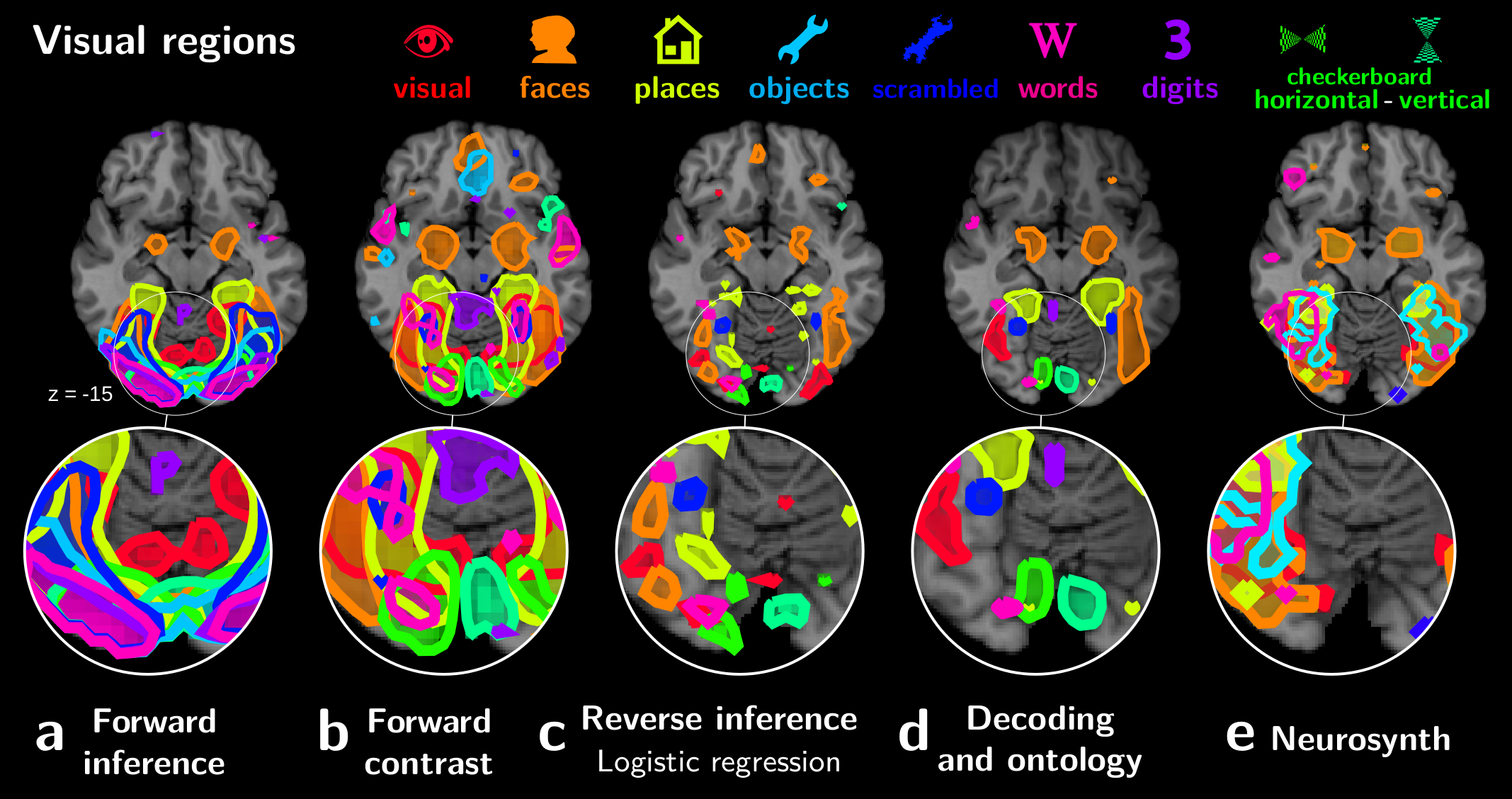
We applied these techniques to the data from 30 different studies, resulting in a detailed break down of the cortex in functionally-specialized modules:
Importantly, the validity of this decomposition in regions is established by the ability of these regions to predict the cognitive aspects of new experimental paradigms.
Predictive models avoid excessive reductionism in cognitive neuroimaging
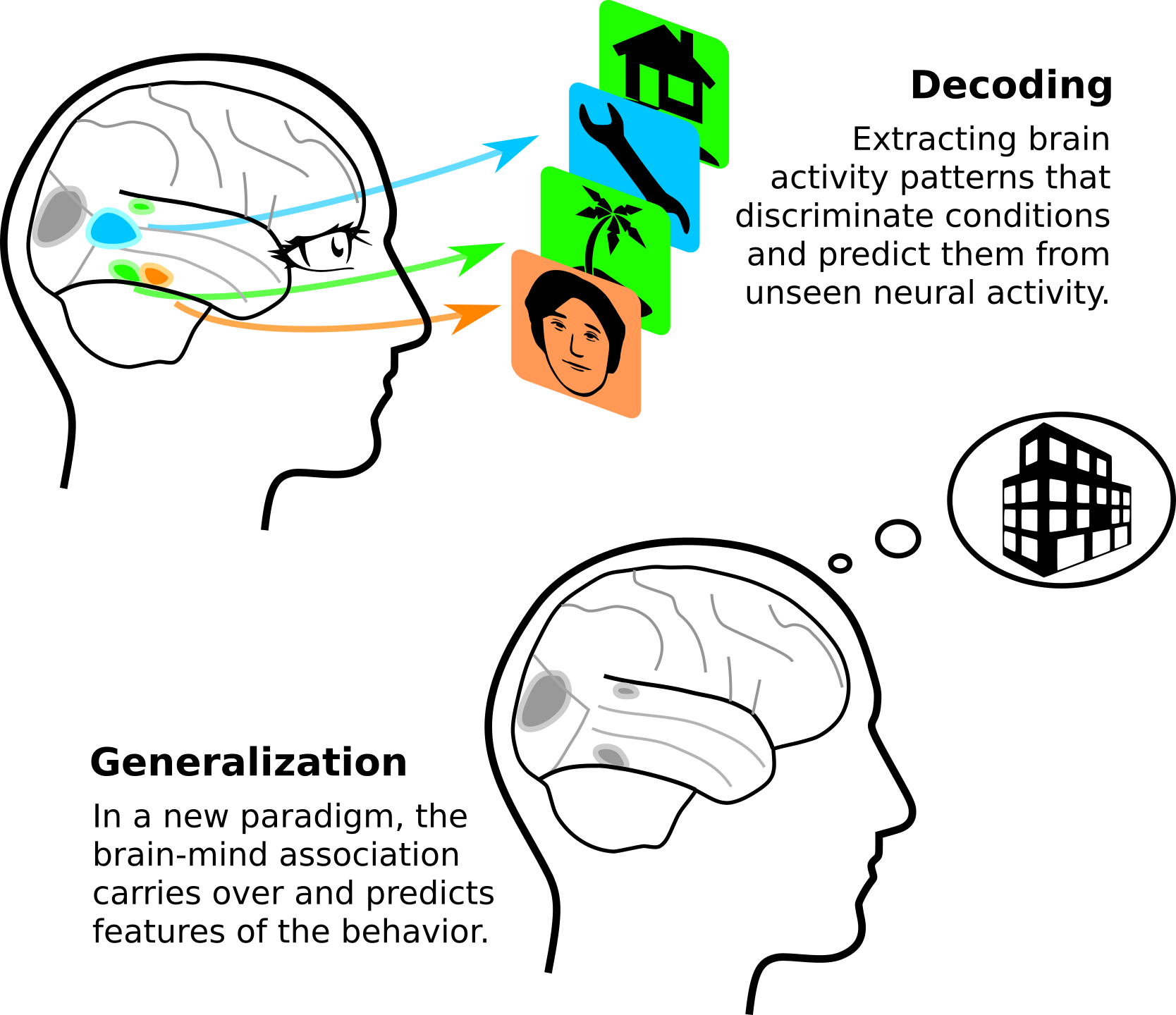
While machine learning is generally seen as an engineering tool to build predictive models or automate tasks, I see in it a central method of modern science. Indeed, it can distill evidence that generalizes from vast –high dimensional– and ill-structured experimental data. Beyond prediction, it can guide understanding.
With Russ Poldrack, we wrote an opinion paper [Varoquaux & Poldrack, Curr Opinion Neurobio 2019] that details why predictive models are important tools to building wider theories of brain function. It reviews many exciting progresses in uncovering with predictive models how brain mechanisms support the mind. It makes the point that ability generalize is a fundamentally desirable priority of scientific inference. Models that are grounded on explicit generalization give a solid path to build broad theories of the mind. Particularly interesting is generalization to significantly different settings, ie going further than typical cross-validation experiments of machine learning, where identical data are artificially split.
Something that is dear to my heart is that we are aiming for quantitative generalization, while psychology often contents itself with qualitative generalization.
Individual Brain Charting, a high-resolution fMRI dataset for cognitive mapping
We are convinced about the importance of analyzing brain response across multiple paradigms, to build models of brain function that generalize across these paradigms. However, addressing such a research program by aggregating multiple studies is hindered by data heterogeneity, due to inter-individual differences or to differing scanners.
Hence, my team, Parietal, has undertook a major data acquisition, the Individual Brain Charting project: scanning a few individuals under a huge amount of cognitive tasks. The data acquisition will last for many years, as the individuals come back to the lab for new acquisitions. The images are of excellent quality, thanks to the unique expertise of our scanning site, Neurospin, a brain-imaging research facility.
The data are completely openly accessible: the raw data, preprocessed data, statistical outputs, alongside with the processing script. We are releasing new data as the project moves forward. This year, we published the data paper [Pinho et al, Scientific Data 2018].
Data accumulation in brain imaging
We are living exciting times, as there are more and more large volumes of shared brain imaging data. OpenfMRI aggregates data in a consistent way across brain-imaging studies. Large projects such as the Human Connectome Project, our Individual Brain Charting project, or the UK BioBank, are designed from the beginning to be shared. We are entering an era of brain-image analysis on many terabytes of data, with dozens of thousands of subjects, compounding hundreds of different clinical or cognitive conditions.
Massive data accumulation opens exciting new scientific prospects, and raises new engineering challenges. Some of these challenges are to scale up neuroimaging data-processing practices, eg inter-subject alignments at the scale of many thousands subjects. Some of these challenges are new to neuroimaging: when compounding hundreds of sources of data into an analysis, the human cost of data integration becomes a major roadblock. As I have become convinced that analysing more, and more diverse, data is an important way forward, I have started working on data intergration per se.
Data science without data cleaning
A new personal research agenda: DirtyData
Challenges to integrating data in a statistical analysis are ubiquitous, including in brain imaging. Data cleaning is recognized as the number one time sink for data scientists. When advising scikit-learn users, including very large companies, I often find that the major roadblock is going from the raw data sources to the data matrix that is input to scikit-learn.
A year ago, I started a new research focus, around the DirtyData project. We now have a team with multiple exciting collaborations, and funding. Our goal is to facilitate statistical analysis of non-curated data. We hope to foster better understanding of how powerful machine-learning models can cope with imperfect, non homogeneous data. As we go, we will publish this understanding, but also distribute code with new methods, and hopefully influence common data-science practices and software. This is an exciting adventure (and yes, we are hiring; see our job offers or contact me).
The topics are vast, at the intersection between database research and statistics. In particular, it calls for integrating machine learning with:
- Knowledge representation
- Information retrieval
- Information extraction
- Statistics with missing data
Similarity encoding: analysis with non-normalized string categories
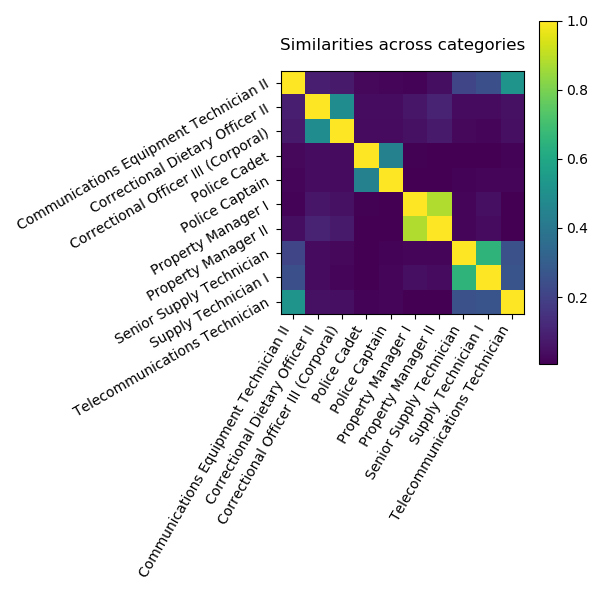
While the DirtyData project is young, we already made progress for analysis of dirty categories, ie categorical data represented with strings that lack curation. These can have typos or other simple morphological variants (eg “patient” vs “patients”), or they can have more structured and fundamental differences, eg arising from the merge of multiple data sources. This latter problem is well-known of database research, where it is seen as a record linkage or alignment problem.
For statistical analysis, in particular machine learning, the problem with these non-curated string categories is that they must be encoded to numerical representations, and classic categorical encodings are not well suited for them. For instance, one-hot encoding leads to very high cardinality.
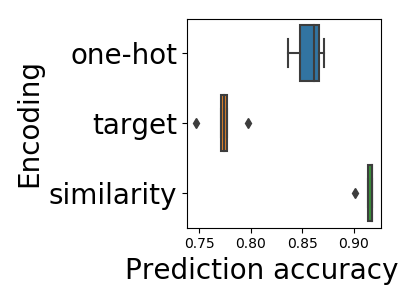
In Cerda et al (2018), we contribute a simple encoding approach, similarity encoding, based on interpolating one-hot encoding with string similarities between the categories.
We ran an extensive empirical study, and show that similarity encoding leads to better prediction accuracy without curation of the data, outperforming all the other approaches that we tried. The paper is purely empirical, but stay tuned: a theoretical analysis of why this is the case is coming soon.
For the benefit of data scientists and researchers, we are released a small Python package, dirty-cat, for learning with dirty categories.
This is just the beginning of the DirtyData project, more exciting work is under way.
Scikit-learn: growth and consolidation

In 2018, a lot of my energy went to consolidating scikit-learn as a project. Describing the work in detail is for another post. However, my main efforts where around growing the team and working on sustainability.
- We established a scikit-learn foundation at Inria, in which companies partner with us to fund scikit-learn development. This took a lot of effort to establish good partnerships and create the legal vessels. Indeed, we want to make sure that the common effort is invested to make scikit-learn better. For instance, working with Intel, who are somewhat running an arms race for computing speed, we improved our test suite, and are slowly but surely learning how to improve our speed.
- A consequence of the foundation is that we are hiring to grow the team (check out our open positions). In 2018, my own team grew, with more excellent people working on scikit-learn, but also joblib, and even contributing to core Python and numpy to improve parallel computing and pickling.
- As the scikit-learn community is growing, it seemed important to formalize a bit more how decisions are made. To me, an important aspect was laying out clearly that the project is still governed by the community, and not partners or people paid by the foundation. We have a draft of a governance document, that is pretty much ready for merge. We also worked on a roadmap. It is a non binding document, but it still was an interesting exercise.
- Scikit-learn 0.20 was released, with many enhancements. And the 0.20 release was followed by two minor releases, to make sure that our users got robust code with backward compatibility.
We are busy finishing a few very interesting studies; next year will be exciting! I hope that we will have much to say about population analysis with brain imaging, which is a amazingly interesting subject.