A couple of weeks ago, we had in Paris the second international nilearn sprint, dedicated to making machine learning in neuroimaging easier and more powerful.
It was such a fantastic experience, as nilearn is really shaping up as a simple yet powerful tool, and there is a lot of enthusiasm. For me, this sprint is a turning point, as I could see people other than the original core team (that spanned out of our research team) excited about the project’s future. Thank you to all who came:
- Ahmed Kanaan
- Andres Hoyos Idrobo
- Alexandre Abraham
- Arthur Mensch
- Ben Cipolli (remote)
- Bertrand Thirion
- Chris Filo Gorgolewski
- Danilo Bzdok
- Elvis Dohmatob
- Julia Hutenburg
- Kamalaker Dadi
- Loic Esteve
- Martin Perez
- Michael Hanke
- Oscar Nájera, working on sphinx-gallery

The sprint was a joint sprint with the MNE-Python team, that makes MEG processing awesome. We also need to thank Alex Gramfort, who did most of the work to set up the sprint, as well as NeuroSaclay for funding, and La paillasse, Telecom, and INRIA for hosting.
Highlights of the sprints results
Plotting of multiple maps
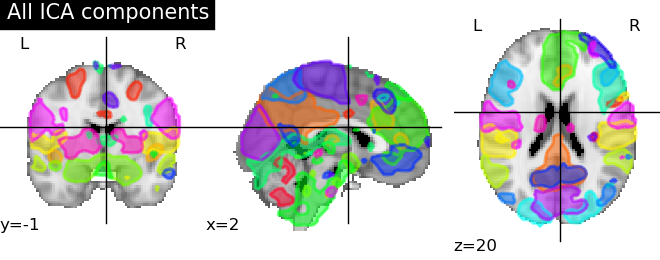
A function to visualize overlays of various maps, eg for a probabilistic atlas, with defaults that try to adapt to the number of maps (see the example). It’s very useful for example for easy visualizing of ICA components.
Sign of activation in glass brain
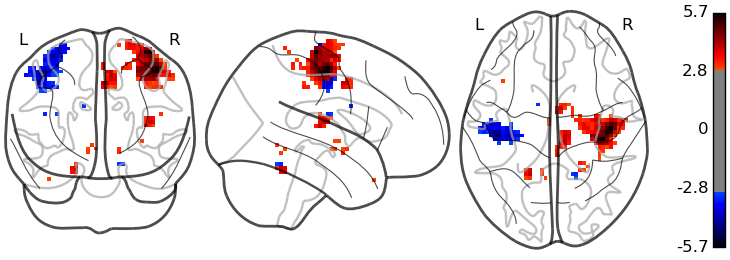
Our glass brain plotting was greatly improved adding amongst other things the option to capture the sign of the activation in the color (see this example).
Spatially-regularized decoder
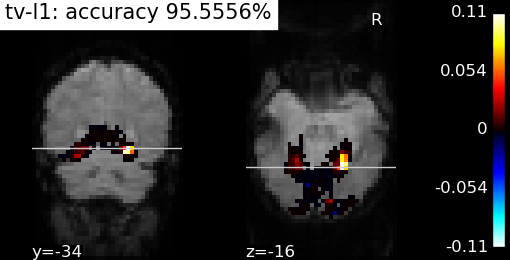
Decoders based on GraphNet and total variation have finally landed in nilearn. This has required a lot of work to get fast convergence and robust parameter selection. At the end of the day, it is much slower than an SVM, but the maps look splendid (see this example).
Sparse dictionary learning
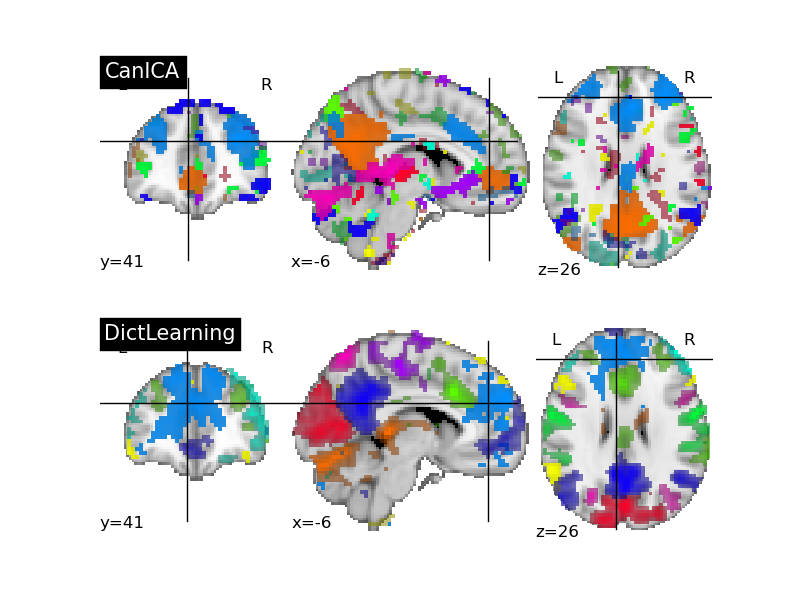
We have almost merged sparse dictionnary learning as a alternative to ICA. Experience shows that on resting-state data, it gives more contrasted segmentation of networks (see this example).
New installation docs
New webpage layout using tabs to display only the installation instruction relevant to the OS of the user (see here). The results are more compact and more clear instructions, that I hope will make our users’ life easier.
CircleCI integration
We now use CircleCI to run the examples and build the docs. This is challenging because our examples are real cases of neuroimaging data analysis, and thus require heavy datasets and computing horse power.
Neurodebian packaging
There are now neurodebian packages for nilearn.
And much more!
Warning
Features listed above are not in the released version of nilearn. You need to wait a month or so.
